How to compute simplicial homology in Python
mathematics
We will consider homology with real coefficients. If you’re already familiar with the subject, you know that some important topological information is lost by doing so, as homology with coefficients in a field ignores torsion effects. It is however simpler to understand. I may write a follow-up article to show how to compute homology with integer coefficients at some point. Most of what I’m going to discuss comes from this excellent lecture on computational algebraic topology by Michael Robinson, who works as an associate professor at the American University. This awesome guy also has a Github repo with computational homology implementations. I’m pretty sure his stuff runs faster than mine, but I wanted to implement something a bit more readable.
Disclaimer: I am not a mathematician, although I very much enjoy mathematics, so I’ll probably be doing some handwaving here and there. If you’re interested in learning computational algebraic topology the right way, find some online lecture like the one I mentioned, or read a reference handbook on the subject (I can recommend [1] which is a very intelligently put and cleverly illustrated overview of applied topology).
Terminology and definitions
We’ll need to give some quick definitions to set the ideas, before we do some code. You can skip this part if you already know what simplicial homology is and you’re eager to get your hands dirty.
Simplicial complex
A simplicial complex can be viewed as a generalization of a graph. It is a set of geometric elements called simplices (singular: simplex) that is closed under inclusion. They can approximate topological spaces (up to homeomorphism), and are used in algebraic topology (along with other kinds of complexes) to make certain calculations possible.
- If
- If
In what follows, if we don’t explicitly specify the codimension, we will assume that it is 1. So a face of a k-simplex
Now all the terminology here is borrowed from geometry for a reason: any abstract simplicial complex, which is a combinatorial object in nature, can be mapped to its geometric counterpart embedded in
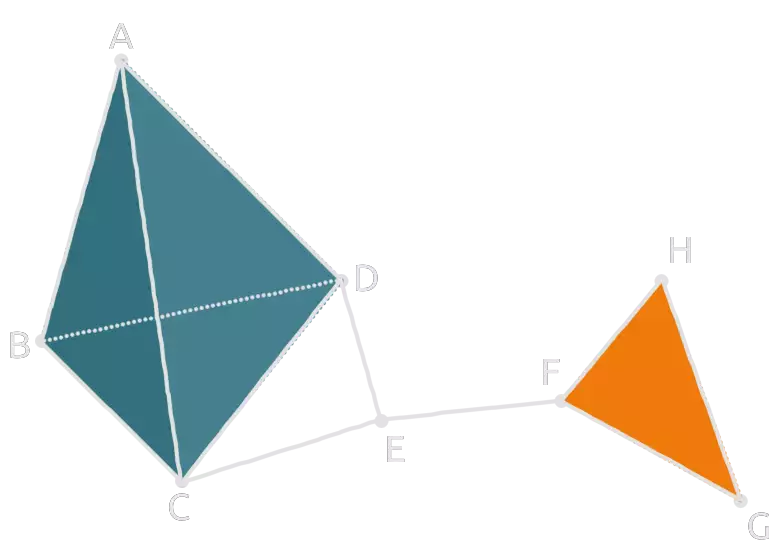
Simplicial complexes have a wide range of applications in data science. Maybe you want to characterize the underlying topology of a communication / information network using something like a Čech complex or a Vietoris-Rips complex (see [5]). Or you’re studying a brain connectome (see [6]). In practice, the simplicial complexes one encounters in research are huge, and computational methods are desired in order to study them. One interesting measurement you can do which gives relevant topological information is simplicial homology.
Simplicial homology is actually a sequence of measurements
Additional structure on simplicial complexes
k-chain spaces
Let
Notice that the dimension of
Without loss of generality, we can consider that abstract simplicial complexes are specified in terms of ordered lists of k-simplices. The order is whatever we like, as long as we are consistent.
Boundary operators
Now we can define the linear maps
So it is sufficient to specify the action of
Here the hat
In
A chain complex
Notice that the collection of k-chains and boundary maps form a graded sequence of the form:
And by
Chain complex
The pattern we identified earlier, where one comes up with a sequence of morphisms between spaces, such that the composition of two consecutive morphisms always gives the zero map, is in fact very recurring in abstract algebra. This setup is so common in fact, that there is a name for it: chain complex.
So any morphism of an exact chain complex will always map a space onto the kernel of the next morphism.
-
-
Homology
Not all chain complexes are exact, that’s a fact. Given an arbitrary chain complex, we would like to quantify how it fails to be exact, and that’s the purpose of homology. In mathematics, when we want to measure by how much a given property fails to be respected by a collection of objects, we usually form a quotient of that collection, in order to mod out the elements that respect this property. We’re left with a quotient space whose elements represent in some sense the many ways to fail respecting this property. I’m not going to properly define quotient spaces here, I assume you already know that much if your goal is to compute homology spaces.
Assuming our
So if
So there is only one coset, therefore the quotient consists of a single element, which means it is trivial. Conversely, we can also show that if all homology spaces are trivial, then the chain complex is exact. So saying that a chain complex is exact or that it has all trivial homology spaces are equivalent statements. This goes to show that homology really quantifies the obstruction to exactness. Moreover, because we have an indexed sequence of homology spaces, it tells us exactly where this obstruction occurs.
Simplicial homology
Let’s go back to our simplicial complex
So we take cycles, and mod out boundaries. The combinatorial interpretation of simplicial homology is somewhat complicated to wrap one’s head around, and I find the geometric interpretation easier to grasp. In the geometric realization, cycles generalize the notion of a cycle in a graph: they represent a sequence of k-dimensional simplices that loop around somehow (and the signs can be interpreted as their respective orientations). And if such a cycle loops around an existing (k+1)-simplex, then it is the boundary of that simplex. But it is possible that the cycle loops around nothing, when its elements are not the faces of a higher dimensional simplex. In this case, we have a hole. The point is that in an arbitrary (abstract) simplicial complex not all cycles are boundaries, and simplicial homology quantifies the obstruction of cycles to be boundaries. So in some sense, it detects holes in the geometric realization.
The dimension of
Computing the boundary operators
In this part, we are going to explicitly compute boundary operators within the context of a toy example, then we’ll derive some Python code to do this automatically.
Manual computation
Because the
A geometric realization would be two triangles sharing an edge, one triangle being filled and the other hollow:
We have one 2-simplex, five edges and four vertices. So the associated chain complex is of the form:
Note that as there is no 3-simplex, numpy library we are going to use can handle these kinds of edge cases seamlessly.
Only
This linear combination of basis vectors of
Here we annotated the rows and columns of our matrix so we can keep track of where simplices are mapped to. Same spiel for
We can check by matrix multiplication that
Implementation
Suppose that our abstract simplicial complex is specified as a list of strings. Each string element can be a single character for a vertex, two characters for an edge and so on. Our previous example would look like this:
X = ["A", "B", "C", "D", "AB", "AC", "BC", "BD", "CD", "BCD"]
Note that we don’t need to specify the empty set, the algorithm does not care about it. Let’s write a boundary() function which will end up producing the full list of boundary operators. The first thing we would like to do, is to group the simplices by dimension. During this step we will also make sure that the simplices are in lexicographic order, that’s the ordering convention I used in the example without mentioning it.
def boundary(complex):
maxdim = len(max(complex, key=len))
simplices = [sorted([spx for spx in complex if len(spx)==i]) for i in range(1,maxdim+1)]
So we first look for the maximal simplex dimension. The max() function is used with parameter key=len to find the string of maximal length within the input array. Then for each possible string length, we form sublists of simplices of the same length. The sorted() function ensures a lexicographic ordering of these simplices within each sublist. If we were to print out the simplices list for X at this stage, we would get:
[['A', 'B', 'C', 'D'], ['AB', 'AC', 'BC', 'BD', 'CD'], ['BCD']]
These sublists represent the respective bases of all the get_faces() that we’ll specify later on. Now we just need to check which simplex
def boundary(complex):
maxdim = len(max(complex, key=len))
simplices = [sorted([spx for spx in complex if len(spx)==i]) for i in range(1,maxdim+1)]
# Iterate over consecutive groups (dim k and k+1)
bnd = []
for spx_k, spx_kp1 in zip(simplices, simplices[1:]):
mtx = []
for sigma in spx_kp1:
faces = get_faces(sigma)
mtx.append([get_coeff(spx, faces) for spx in spx_k])
bnd.append(np.array(mtx).T)
return bnd
The list comprehension [get_coeff(spx, faces) for spx in spx_k] will form a row vector containing the coefficients mtx for all mtx before we push it in the bnd list.
The helper functions are defined as follows:
def get_faces(lst):
return [lst[:i] + lst[i+1:] for i in range(len(lst))]
def get_coeff(simplex, faces):
if simplex in faces:
idx = faces.index(simplex)
return 1 if idx%2==0 else -1
else:
return 0
The function get_faces() takes a string input and returns all the possible substrings obtained by deleting a character at index get_coeff() takes a simplex and a list of faces, and returns either the index parity if the input simplex is found in the faces list, or zero otherwise.
We can check our boundary() function against the example abstract simplicial complex X and print out the boundary operators it returns:
print(boundary(X))
[array([[-1, -1, 0, 0, 0],
[ 1, 0, -1, -1, 0],
[ 0, 1, 1, 0, -1],
[ 0, 0, 0, 1, 1]]),
array([[ 0],
[ 0],
[ 1],
[-1],
[ 1]])]
We recognize the matrices of
Aside: Computing (co)kernels and (co)images
For what comes next, we will need to be able to compute numerically the bases of the kernels and cokernels of arbitrary matrices. For those who read a few articles on this blog, you’ll know by now that we love singular value decomposition, like, a lot. It turns out, once again, they’re the tool for the job. Let’s write a quick definition, and recapitulate what an SVD does.
Singular values and Singular Value Decomposition
- The columns
- The columns
Then,
- The vectors
- Similarly, the vectors
- Note that the cokernel and image are in direct sum:
- And so are the kernel and coimage:
Assuming descending order of the singular values, we have that
We’re only going to need
Implementation
So to find a basis for the kernel of a matrix, according to np.linalg.svd(), and extract the rows of
def kernel(A, tol=1e-5):
_, s, vh = np.linalg.svd(A)
singular = np.zeros(vh.shape[0], dtype=float)
singular[:s.size] = s
null_space = np.compress(singular <= tol, vh, axis=0)
return null_space.T
The array s returned by np.linalg.svd() is one-dimensional, and contains only the singular values big enough so their floating point representation is non-zero. But it could happen that some of these should actually be interpreted as null singular values. So we put them in a bigger array singular of size the number of rows of vh, padding with zeros, and we use the np.compress() function to extract the rows of vh where the values of singular are close enough to zero (using an input tolerance parameter). The output of np.compress() is a matrix whose rows are the ones we selected. We transpose this matrix before we return it, so in the end this function outputs a matrix whose columns span the kernel of the input matrix. You’ll see later on why we do this, rather than returning a list of vectors.
As for the cokernel, we do a similar manipulation following our previous result
def cokernel(A, tol=1e-5):
u, s, _ = np.linalg.svd(A)
singular = np.zeros(u.shape[1], dtype=float)
singular[:s.size] = s
return np.compress(singular <= tol, u, axis=1)
The code above is more or less a copy paste of this implementation by Michael Robinson and Chris Capraro. I did come up with something similar on my own, but I found this version more elegant as Python code goes.
Computing homology
There are multiple algorithms out there to compute homology. I should mention the great article [7] (do check out this guy’s blog) which shows a way to compute homology dimensions directly by simultaneous row reduction on consecutive boundary operators and pivot counting.
What we will do instead, is to instantiate
The endofunctor trick
This method was presented by Michael Robinson in his lecture on computational algebraic topology on Youtube, and it’s gold. Let’s consider the chain complex around
There is a natural inclusion
To see why the cokernel of
A few practical observations concerning these new maps:
- Because
np.linalg.lstsq()function. - The product
Because our previously defined kernel() and cokernel() functions cast the respective basis vectors in transition matrix form, they can be used to compute the matrices of
Implementation
Recall that from our previous boundary() function, we get a list of matrices, one for each boundary operator. We now define a homology() function that takes in this list of matrices, and outputs a list of homology spaces specified by their respective bases. The list of boundary operators shall be augmented by two additional zero maps of the appropriate size (I mentioned that numpy can handle zero-size matrices). We will iterate over all consecutive pairs of boundary operators in the augmented list, and each time compute the product
def homology(boundary_ops, tol=1e-5):
# Insert zero maps
mm = boundary_ops[-1].shape[1]
nn = boundary_ops[0].shape[0]
boundary_ops.insert(0, np.ones(shape=(0, nn)))
boundary_ops.append(np.ones(shape=(mm, 0)))
H = []
for del_k, del_kp1 in zip(boundary_ops, boundary_ops[1:]):
kappa = kernel(del_k, tol)
# Solve for psi
psi, _, _, _ = np.linalg.lstsq(kappa, del_kp1, rcond=None)
# Compute homology
ksi = cokernel(psi, tol)
H.append(np.dot(kappa, ksi))
return H
To be a bit provocative here, in the end, computing a homology takes four lines of code.
Computing the Betti numbers
Once we have the bases of each
def betti(H):
return [basis.shape[1] for basis in H]
These are known as the Betti numbers
Putting it all together
Let’s now write a proper main() function that will read a Json file containing various simplicial complexes accompanied by a short description, so we can experiment with this a bit more easily:
def main():
with open('./complexes.json') as data_file:
data = json.load(data_file)
for attr, obj in data.items():
print(f'[{attr}]')
print(obj['description'])
bnd = boundary(obj['complex'])
H = homology(bnd)
b = betti(H)
print(f'Betti numbers: {b}')
And here’s how the example simplicial complex is encoded in the Json file:
{
"C1": {
"description": "Two triangles sharing an edge, one of them being hollow, the other filled",
"complex": ["A", "B", "C", "D", "AB", "AC", "BC", "BD", "CD", "BCD"]
}
}
Running the main() function should now output:
[C1]
Two triangles sharing an edge, one of them being hollow, the other filled
Betti numbers: [1, 1, 0]
That’s it! One connected component, a single 1-hole (the hollow triangle), and no 2-hole. And for the grand finale, we are going to compute the homology of a 2-torus. [2] mentions an efficient triangulation of a two dimensional torus, from which I could build a simplicial complex:
Here is the Json data:
{
"T2": {
"description": "A triangulated 2-torus",
"complex": ["A","B","C","D","E","F","G",
"AB","AC","AD","AE","AF","AG",
"BC","BD","BE","BF","BG",
"CD","CE","CF","CG",
"DE","DF","DG",
"EF","EG",
"FG",
"ABD","ABF","ACD","ACG","AEF","AEG",
"BCE","BCG","BDE","BFG",
"CDF","CEF",
"DEG","DFG"]
}
}
And our program now outputs:
[T2]
A triangulated 2-torus
Betti numbers: [1, 2, 1]
Which is precisely the Betti numbers for the 2-torus: one connected component, two circular 1-holes, and one 2-hole (the volume inside the torus). Imagine doing that by hand…
Conclusion
Simplicial complexes appear naturally when one forms the Vietoris-Rips complex of a point cloud in
Acknowledgements
Many thanks to nervous_lemma for the proofreading help!
Sources
- [1] R. Ghrist, “Elementary Applied Topology”, ed. 1.0, Createspace, 2014
- [2] J. Michael Boardman, “The Torus Triangulated”
- [3] Roman, Steven. Advanced Linear Algebra. 3rd ed, Springer, 2007, p.444
- [4] Zomorodian, A., Carlsson, G. Computing Persistent Homology. Discrete Comput Geom 33, 249–274 (2005).
- [5] M. E. Aktas et al. Persistence homology of networks: methods and applications. Applied Network Science (2019).
- [6] Giusti, C., Ghrist, R. & Bassett, D.S. Two’s company, three (or more) is a simplex. J Comput Neurosci 41, 1–14 (2016).
- [7] j2kun, Computing Homology
 Orange Wire
Orange Wire





